Using DNA science to monitor invasive and endangered species
Environmental DNA analysis and the use of emerging biological technologies provide a new approach to monitoring for invasive and endangered species, detecting pathogens, characterizing biodiversity, and assessing aquatic ecosystems.
By Yuwei Xie, Chris DeBeer, and John Giesy

Lakes, rivers, and other bodies of water are facing increasing pressures from climate and environmental change, human development, agriculture and industry, and invasive species. Aquatic ecosystems are under threat and regulators, scientists, and local communities face significant challenges in protecting these ecosystems. This requires reliable and rapid detection of changes in environments and prediction of future trends to be able to maintain and enhance sustainability of ecosystems.
Researchers at the University of Saskatchewan and the University of Waterloo have been leading a Global Water Futures project called Next Generation Solutions to Ensure Healthy Water Resources for Future Generations. This project develops and applies emerging and transformative technologies in biology and chemistry, DNA science, and bioinformatics to monitor current status and predict future trends of aquatic ecosystems exposed to stressors. Together, these transformative technologies have potential to provide more rapid, comprehensive, and objective assessments of ecosystem status, including presence of rare and endangered species, in near real time, while reducing costs and the need for taxonomic expertise.
Environmental DNA as a new tool for ecosystem monitoring
All organisms leave their DNA—the molecules that contain their genetic blueprints and are unique to each organism—in the ecosystem as they shed cells, tissues, and wastes, or die. RNA, which is structurally similar to DNA but often contains only a single stand, is also left behind. DNA and RNA can then be extracted from samples collected in lakes, rivers, or sediment, and identified in the laboratory. Environmental DNA (or eDNA) analysis can deliver a high level of taxonomic specificity without the need for labor-intensive collections and identifications of individual organisms. Further, these technologies are capable of detecting as little as a single molecule of DNA or RNA. Until recently, however, the application of eDNA analysis was limited by low-throughput sequencing (the way to determine the order of nucleotides along a strand of DNA). Now, high-throughput sequencing, also known as next-generation sequencing (NGS) technology, is capable of sequencing hundreds of millions of DNA molecules at a time.
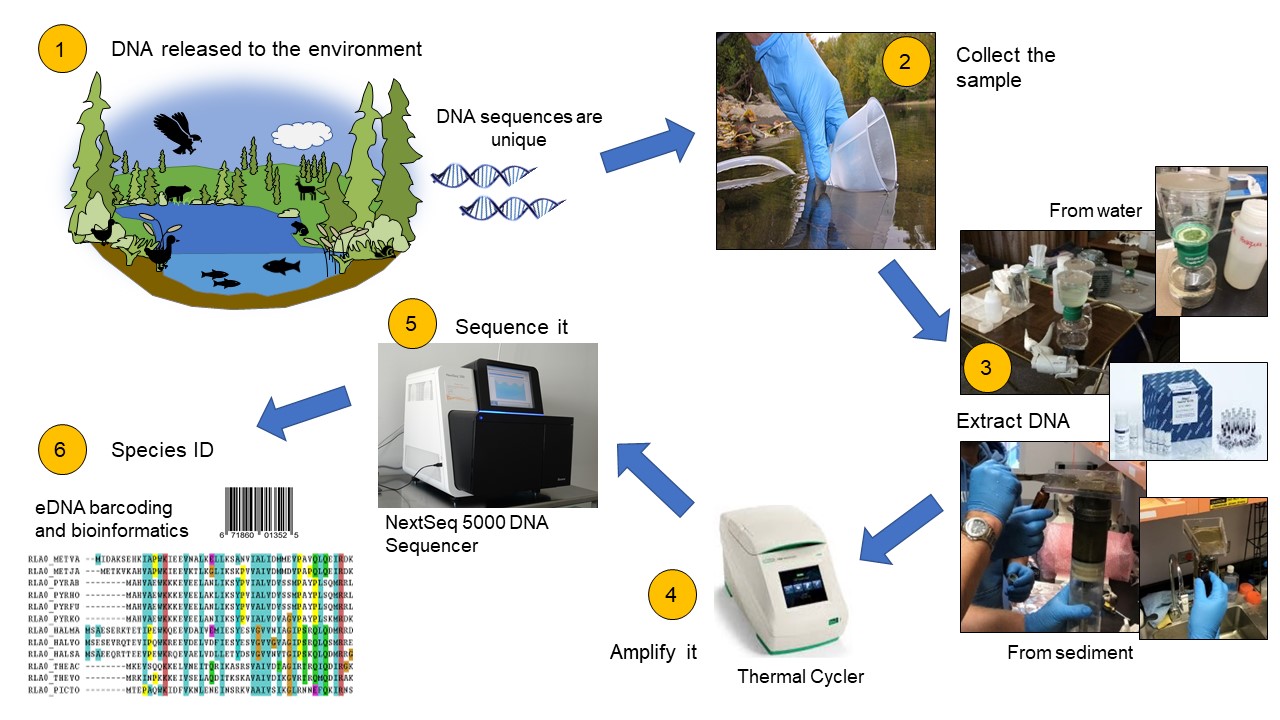
Schematic depicting the processes involved in ecosystem assessment using eDNA. Newly developed standard operating procedures for each step can be found at https://gwf.usask.ca/edna/resources/standard-operating-procedures.php.
Organisms shed their DNA onto the land and into streams, rivers, and lakes. eDNA can originate from any tissue or biological material left behind by an organism such as cells, tissues, urine, hair, skin, mucus, and dead individuals leaking genetic material. Rates of shedding can vary depending on factors such as species and size, life stage, and organism health.
eDNA can be obtained by collecting environmental samples such as water, sediment, soil, air, etc., without the source organisms having to be visually present. Samples are collected using careful, standardized procedures—essential to prevent contamination of the samples with DNA from other sources.
In the lab, eDNA is extracted, purified, and quantified from environmental samples using a rigorous and quality-controlled set of procedures. This generates an isolated eDNA sample that is preserved and stored for further analysis.
Target DNA is amplified to generate a sufficient amount of the genetic material for sequencing analysis in the next step. This involves a process in which DNA molecules are copied, creating replicates with the same genetic sequence as the original.
DNA sequencing is performed to determine the order of nucleotides (the molecular units of DNA forming a three-dimensional chain). Next-generation technology, such as the Illumina sequence-by-synthesis sequencer (shown), are capable of sequencing multiple DNA molecules in parallel.
Determination of species presence and abundance are made through advanced bioinformatics tools (for example, taxonomic annotation). These use computer software to characterize complex and large datasets of DNA sequences.
Because DNA breaks down more slowly and persists longer in an ecosystem than RNA, eDNA provides a surrogate for the status of an ecosystem over a period of time, while eRNA is more like a snapshot of its status at a given time.
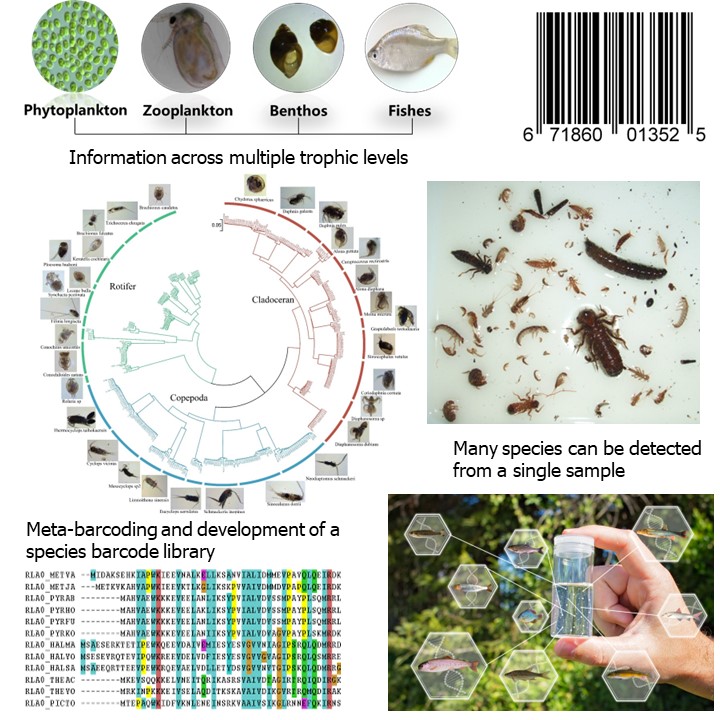
The Next Generation Solutions research team has been focusing on making this technology functional in a way so that the data collected can be compared and interpreted in the context of information collected historically based on traditional taxonomic nomenclature. This involves establishment of libraries of DNA/RNA sequences of what are known as operational taxonomic units, similar to a barcode database for different species that can be annotated with Latin species names. Referred to as meta-barcoding, this allows for the simultaneous identification of many species from a single sample. The project has established operational taxonomic units of Canadian freshwater fish, macrobenthos, and zooplankton.
Strengths of eDNA- and eRNA-based monitoring and assessment include:
- reduced field survey time,
- little or no impact on ecosystems (non-invasive),
- species identification from DNA sequences is often easier and more accurate than identification by observation of external morphology,
- many aquatic species may be detected from a single water sample.
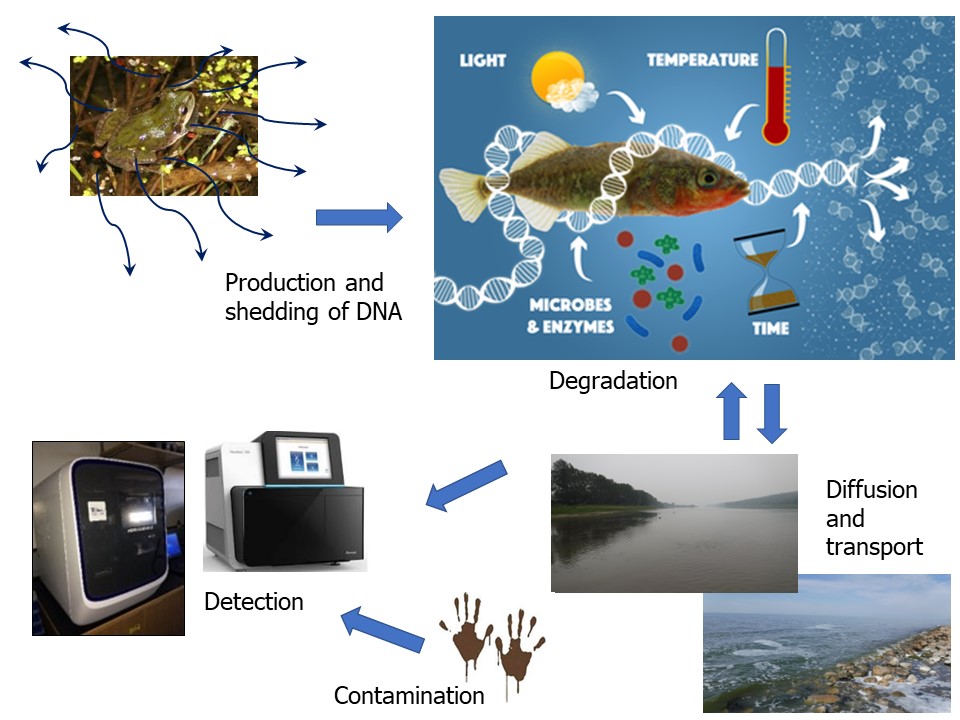
There are challenges affecting the detection of eDNA. Rates of shedding by aquatic micro- and macro-organisms can vary depending on factors such as species and size, life stage, and organism health. Environmental and hydrological factors, such as temperature, rates of flow of water, acidity, ultraviolet light intensity and salinity can affect persistence of DNA and RNA in water.
The research has been focusing on assessing rates, reliability, and variability of detection of eDNA/eRNA for many key species of concern. Results have shown very good comparison between field survey information and eDNA/eRNA sequences, providing accurate indicators for species presence, abundance, and biomass at sites across Canada and in several other countries around the world. Through this work, the project has advanced the reliability of eDNA platforms for enhancing ecological monitoring and risk assessment of freshwater ecosystems.
The work has been described in detail and results have been published in several scientific journals, listed and available here:
Kim, T., Lee, C., Lee, J., Bae, H., Noh, J., Hong, S., Kwon, B.O., Kim, J.J., Yim, U.H., Chang, G.S. and Giesy, J.P., 2022. Best Available Technique for the Recovery of Marine Benthic Communities in a Gravel Shore after the Oil Spill: A Mesocosm-based Sediment Triad Assessment. Journal of Hazardous Materials, p.128945.
Fuzzen, M.L., Harper, N.B., Dhiyebi, H.A., Srikanthan, N., Hayat, S., Peterson, S.W., Yang, I., Sun, J.X., Edwards, E.A., Giesy, J.P. and Mangat, C.S., 2022. Multiplex RT-qPCR assay (N200) to detect and estimate prevalence of multiple SARS-CoV-2 Variants of Concern in wastewater. medRxiv.
Ankley, P.J., Graves, S.D., Xie, Y., DeBofsky, A., Weber, A., Brinkmann, M., Palace, V.P., Liber, K., Hecker, M., Janz, D.M. and Giesy, J.P., 2022. Effects of in situ experimental selenium exposure on finescale dace (Phoxinus neogaeus) gut microbiome. Environmental Research, 212, p.113151.
Xie, Y., Challis, J.K., Oloye, F.F., Asadi, M., Cantin, J., Brinkmann, M., McPhedran, K.N., Hogan, N., Sadowski, M., Jones, P.D. and Landgraff, C., 2022. RNA in Municipal Wastewater Reveals Magnitudes of COVID-19 Outbreaks across Four Waves Driven by SARS-CoV-2 Variants of Concern. ACS ES&T Water.
Weber, A., Xie, Y., Challis, J.K., DeBofsky, A., Ankley, P.J., Hecker, M., Jones, P. and Giesy, J.P., 2022. Effects of aqueous fluoxetine exposure on gut microbiome of adult Pimephales promelas. Science of the Total Environment, 813, p.152422.
Hu, D., Giesy, J.P., Guo, M., Ung, W.K., Kong, Y., Mok, K.M. and Lee, S.M.Y., 2021. Temporal Patterns of Bacterial and Viral Communities during Algae Blooms of a Reservoir in Macau. Toxins, 13(12), p.894.
DeBofsky, A., Xie, Y., Challis, J.K., Ankley, P.J., Brinkmann, M., Jones, P.D. and Giesy, J.P., 2022. 16S rRNA metabarcoding unearths responses of rare gut microbiome of fathead minnows exposed to benzo [a] pyrene. Science of The Total Environment, 807, p.151060.
Ankley, P.J., Xie, Y., Havens, S., Peters, L., Timlick, L., Rodriguez-Gil, J.L., Giesy, J.P. and Palace, V.P., 2022. RNA metabarcoding helps reveal zooplankton community response to environmental stressors. Environmental Pollution, 292, p.118446.
Wang, X., Wu, F., Zhao, X., Zhang, X., Wang, J., Niu, L., Liang, W., Leung, K.M.Y. and Giesy, J.P., 2022. Enlightenment from the COVID-19 pandemic: the roles of environmental factors in future public health emergency response. Engineering, 8, pp.108-115.
Mu, Y., Shao, M., Zhong, B., Zhao, Y., Leung, K.M., Giesy, J.P., Ma, J., Wu, F. and Zeng, F., 2021. Transmission of SARS-CoV-2 virus and ambient temperature: a critical review. Environmental Science and Pollution Research, 28(28), pp.37051-37059.
Ankley, P.J., Xie, Y., Black, T.A., DeBofsky, A., Perry, M., Paterson, M.J., Hanson, M., Higgins, S., Giesy, J.P. and Palace, V., 2021. Using zooplankton metabarcoding to assess the efficacy of different techniques to clean-up an oil-spill in a boreal lake. Aquatic Toxicology, 236, p.105847.
Chik, A.H., Glier, M.B., Servos, M., Mangat, C.S., Pang, X.L., Qiu, Y., D'Aoust, P.M., Burnet, J.B., Delatolla, R., Dorner, S. and Geng, Q., 2021. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada. Journal of Environmental Sciences, 107, pp.218-229.
DeBofsky, A., Xie, Y., Challis, J.K., Jain, N., Brinkmann, M., Jones, P.D. and Giesy, J.P., 2021. Responses of juvenile fathead minnow (Pimephales promelas) gut microbiome to a chronic dietary exposure of benzo [a] pyrene. Environmental Pollution, 278, p.116821.
Wang, Y., Chen, K., Gao, J., Wang, M., Dong, J., Xie, Y., Giesy, J.P., Jin, X. and Wang, B., 2021. Environmental DNA of preservative ethanol performed better than water samples in detecting macroinvertebrate diversity using metabarcoding. Diversity and Distributions, 27(10), pp.1989-2002.
DeBofsky, A., Xie, Y., Jardine, T.D., Hill, J.E., Jones, P.D. and Giesy, J.P., 2020. Effects of the husky oil spill on gut microbiota of native fishes in the North Saskatchewan River, Canada. Aquatic Toxicology, 229, p.105658.
Chen, L., Giesy, J.P., Adamovsky, O., Svirčev, Z., Meriluoto, J., Codd, G.A., Mijovic, B., Shi, T., Tuo, X., Li, S.C. and Pan, B.Z., 2021. Challenges of using blooms of Microcystis spp. in animal feeds: A comprehensive review of nutritional, toxicological and microbial health evaluation. Science of The Total Environment, 764, p.142319.
DeBofsky, A., Xie, Y., Grimard, C., Alcaraz, A.J., Brinkmann, M., Hecker, M. and Giesy, J.P., 2020. Differential responses of gut microbiota of male and female fathead minnow (Pimephales promelas) to a short-term environmentally-relevant, aqueous exposure to benzo [a] pyrene. Chemosphere, 252, p.126461.
Lee, A.H., Lee, J., Hong, S., Kwon, B.O., Xie, Y., Giesy, J.P., Zhang, X. and Khim, J.S., 2020. Integrated assessment of west coast of South Korea by use of benthic bacterial community structure as determined by eDNA, concentrations of contaminants, and in vitro bioassays. Environment International, 137, p.105569.
Zhang, X., Tang, S., Wang, M., Sun, W., Xie, Y., Peng, H., Zhong, A., Liu, H., Zhang, X., Yu, H. and Giesy, J.P., 2019. Acid mine drainage affects the diversity and metal resistance gene profile of sediment bacterial community along a river. Chemosphere, 217, pp.790-799.
Tse, T.J., Doig, L.E., Tang, S., Zhang, X., Sun, W., Wiseman, S.B., Feng, C.X., Liu, H., Giesy, J.P., Hecker, M. and Jones, P.D., 2018. Combining high-throughput sequencing of seda DNA and traditional paleolimnological techniques to infer historical trends in cyanobacterial communities. Environmental science & technology, 52(12), pp.6842-6853.
Xie, Y., Zhang, X., Yang, J., Kim, S., Hong, S., Giesy, J.P., Yim, U.H., Shim, W.J., Yu, H. and Khim, J.S., 2018. eDNA-based bioassessment of coastal sediments impacted by an oil spill. Environmental Pollution, 238, pp.739-748.
Zhang, L., Xu, E.G., Li, Y., Liu, H., Vidal-Dorsch, D.E. and Giesy, J.P., 2018. Ecological risks posed by ammonia nitrogen (AN) and un-ionized ammonia (NH3) in seven major river systems of China. Chemosphere, 202, pp.136-144.
Building partnerships and putting the research into practice

The Next Generation Solutions research team have partnered with industries and provincial agencies to demonstrate where eDNA technologies can be used in monitoring and surveillance programs to make them more rapid and increase performance over broader areas for less cost. Collaborative work has been focusing on the comparison of eDNA methodologies to conventional methods at monitoring sites, including boreal lakes, rivers and streams of Saskatchewan, Manitoba, and Ontario, as well as lab experiments.
The work is aimed at developing eDNA ecosystem monitoring solutions for managers and practitioners. Applications include:
- Characterization of the ecological baseline of watersheds in cold regions,
- Determination of the ecological effects of human activities in controlled experiments and monitored sites,
- Comparison of aquatic communities of Canadian lakes, rivers and streams,
- Detection of invasive and threatened species at various sites of interest,
- Monitoring for effects of climate change and eutrophication
Newly developed and validated eDNA tools will create opportunities to better assess and predict changes in the Canadian environment. These emerging technologies have implications for contributing to international effort to improve water quality and promote better management of our aquatic resources globally.
-Lynn P. Weber, Professor & Acting Department Head, Veterinary Biomedical Sciences, University of Saskatchewan

